Bladder cancer variants share aggressive cell state and gene expression features
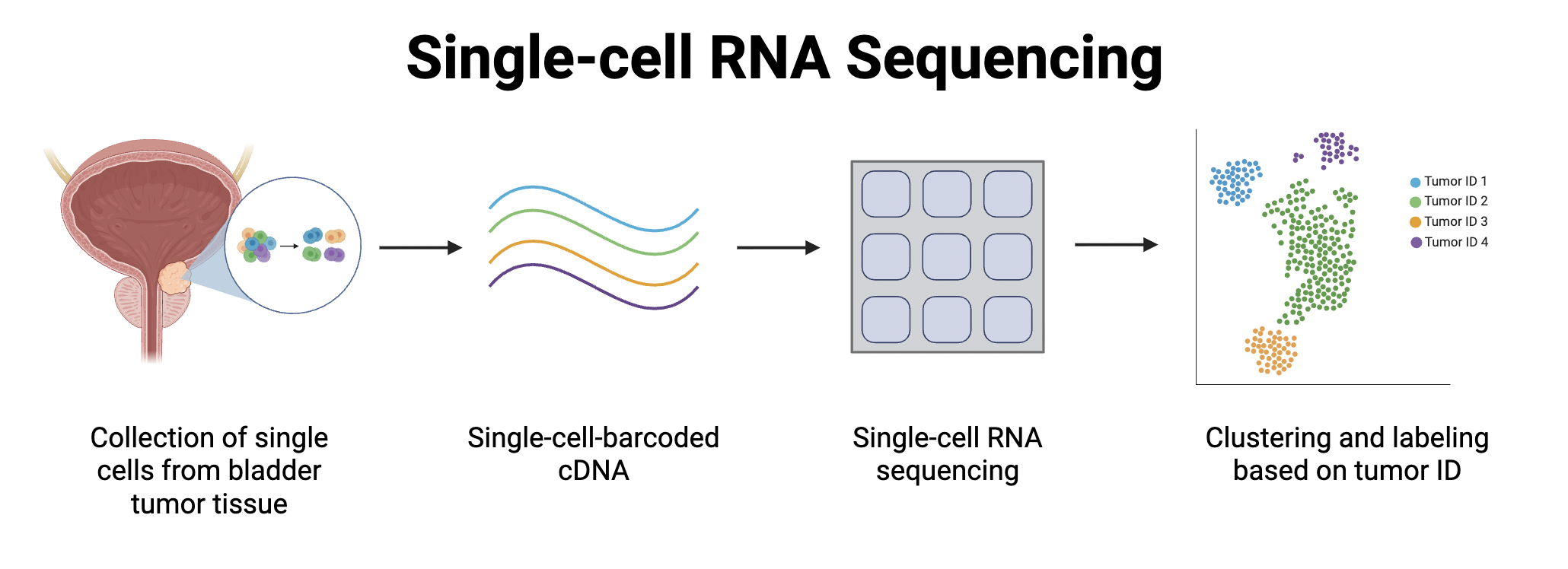
RNA is collected from individual cells of tumor tissue. The RNA is then labelled and sequenced, with sequencing data converted into graphs to visualize similarities and differences in gene expression.
Written by Wendy Xu, Schematic by Lexi Bean
What is the Study About?
Urothelial carcinoma (UC), also known as transitional cell carcinoma, is the most common type of bladder cancer. It begins in the urothelium tissue,which lines the urinary tract, hence the name “urothelial carcinoma." Bladder cancer can have histologic variant (HV) subtypes, or distinct cancer patterns that don’t fall under other broader classifications. They are found more rarely, in up to 25% of all bladder tumors. These subtypes are often more aggressive and more resistant to standard therapy compared to conventional urothelial carcinoma.
Histologic variant subtypes of bladder cancer remain understudied, and much is unknown about the patterns of gene expression that account for their worse clinical outcomes. RIn this study, researchers at the University of California, San Francisco and University of California Santa Barbara, Santa Barbara set out to discover shared characteristics in histological variant subtypes and identify potentially targetable molecular features for treatment.
How Was the Study Conducted?
In this study, tissue samples from 3 regular urothelial carcinomas (UC) and 9 histological variant (HV) tumors were collected and sequenced. Single-cell RNA sequencing (scRNA-seq) was used to determine the RNA composition of these tumors, because it requires relatively smaller sample sizes compared to bulk RNA sequencing.
Using data from the sequencing, the researchers generated a clustered Uniform Manifold Approximation and Projection (UMAP) to visualize and group similar patterns of gene expression. Data points were color coded, and clustered dots indicated genetic similarities between the cells.
What Did They Discover?
On the UMAP they created, researchers found that Cluster 13 contained cancer cells from multiple different HV subtype tumors. This suggests that Cluster 13 represents a tumor cell state that characterizes multiple HV tumors, not just one particular HV subtype. Upon more analysis, they found that the MUC4, MUC17, MUC16, KRT24, and WISP2 genes in Cluster 13 had significantly different expression levels compared to other clusters. Knowing what genes are differentially expressed in HV cells can help researchers understand what “sets them apart.” From there, scientists can then figure out which specific genes may have caused the increased aggressiveness of HV tumors, and what treatments may be more effective against HV tumors.
Researchers also found that Cluster 13 likely represents convergent cell states shared by various HV subtypes. This means that the similar gene expression patterns in Cluster 13 cells developed independently rather than together, indicating potential for a single unified strategy that can treat multiple HV subtypes at once.
The top differentially expressed gene in the HV group was TM4SF1, which is cell surface localized and also implicated in other aggressive bladder cancers[1] as a cell cycle and apoptosis regulator. Since there were no FDA-approved TM4SF1-directed therapeutic agents, researchers used chimeric antigen receptor (CAR) T cell therapy to target bladder cancer lines expressing TM4SF1 in mice. Chimeric antigen receptor T cell therapy uses T cells, which are a type of immune cell that naturally have surface proteins called receptors to target pathogens or abnormal cells. By genetically modifying these T-cells, scientists can make the T-cells produce specific chimeric antigen receptors. In this case, the chimeric antigen receptors specifically targeted TM4SF1. Treatment in the mice proved effective, significantly increasing survival rates compared to untreated mice. Treated mice also had stable weights and no obvious lung damage.
Why Is This Significant?
Histological variants of cancer are poorly understood in part because they are uncommon and unique. ScRNA-seq allowed the researchers to derive insights about HV cancer biology in a relatively small sample of tumors, emphasizing the potential of scRNA-seq technologies in precision cancer medicine research.
Several genes characterize the “Cluster 13” cell state, such as CA125. They can potentially be used as biomarkers, or molecular targets, for therapy. Certain FDA-approved agents such omipalisib and quizartinib were predicted to be more effective against this Cluster 13 compared to other tumor cells.
Researchers discovered that most HV tumors exhibit increased expression of TM4SF1, which has already been implicated in other aggressive bladder cancers and even other cancer cell types. This study demonstrates in mice that TM4SF1 can be targeted by CAR T cells, indicating potential for development of new therapies. This study’s preclinical testing of TM4SF1-CAR T cells lays a foundation for future clinical trials in bladder cancer and also other tumor types expressing TM4SF1.
A link to the publication can be found here: https://pmc.ncbi.nlm.nih.gov/articles/PMC12174346/